Gene deFuser:
Gene deFuser utilizes BLAST to detect similarity between the termini of a protein sequence and the sequences in a database of orthologous protein groups. The ortholog database used in this program is a subset of the eukaryotic COG (clusters of orthologous genes) database. A list of COG identifiers is generated for each end of the protein based on the BLAST results. The list of identifiers found for the N-terminus is then compared with the list for the C-terminus. A typical non-fused protein will match both the beginning and end of one or more COGs, and as a result will return the identifier of the same COG at both the N-terminus and the C-terminus. A protein that returns a matching COG identifier at both ends is presumed to be non-fused and is excluded as a possible fusion gene. Proteins that do not return any COG hit in common at both ends are considered candidate fusion proteins.
Input:
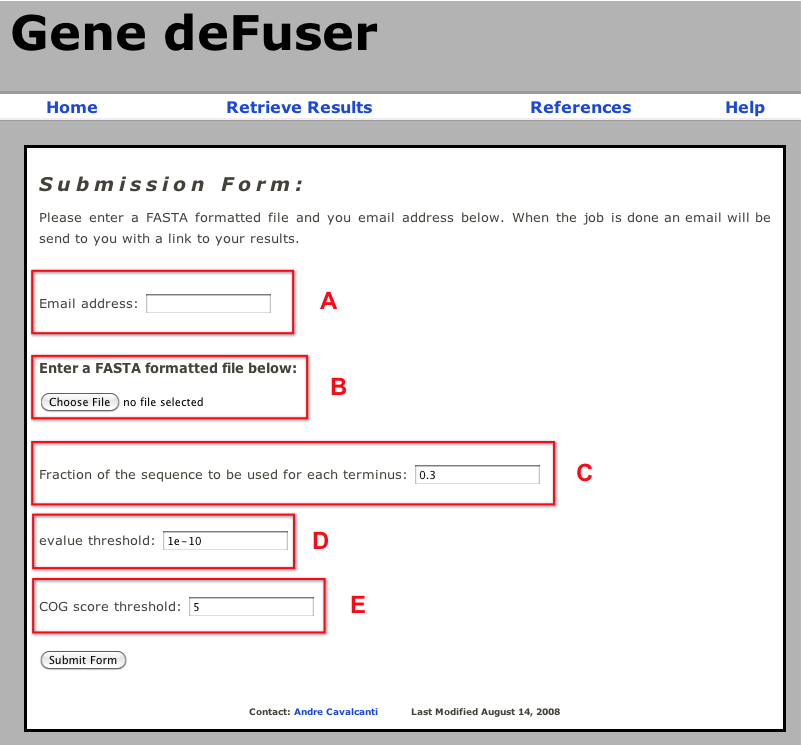
A) Email address to which an email will be sent when the calcualtions are done. Because, depending on the number of proteins to be analyzed, the program can take a few hours to run, the user is required to provide an email address to receive a message indicating the completion of the job. This email will be sent from: genedefuser@gmail.com.
B) FASTA file containing the proteins to be analyzed.
C) Fraction of the sequence to be used for each terminus. Gene deFuser works by BLASTing the N- and C-termini of a protein against a database of orthologous gene groups (COG) and comparing the hits to both ends of the protein. This parameter defines what fraction of the protein sequence will be used as the N- and C-termini queries. For example, if the fraction is 0.3 and the protein is 1000 amino acids long, the first 300 amino acids will be used as the N-terminus and the last 300 amino acids will be used as the C-terminus.
D) Evalue threshold. This parameter determines the maximum e-value for which a hit will be considered in the calculations.
E) COG score threshold. Following the BLAST search of the N- and C- termini of the protein against the COG database a composite score is calculated for each COG that has at least one hit with e-value smaller than the threshold in "D". This composite score is based on the methodology of Zhou and Landweber (2007. Nucleic Acids Res. 35:W678-682). Only COGs with a composite score larger than this threshold will be used in the calcualtions.
Output:
When the program finishes analyzing the user-submitted file an email is sent to the user with the job number. The user can then go to the "Retrieve Results" page and check the results.
The main results page contains a link to download all the results as a .zip file and a list of all the candidate fusion proteins found by the program.
Clicking on a protein in the list will take the user to a candidate fusion gene page.
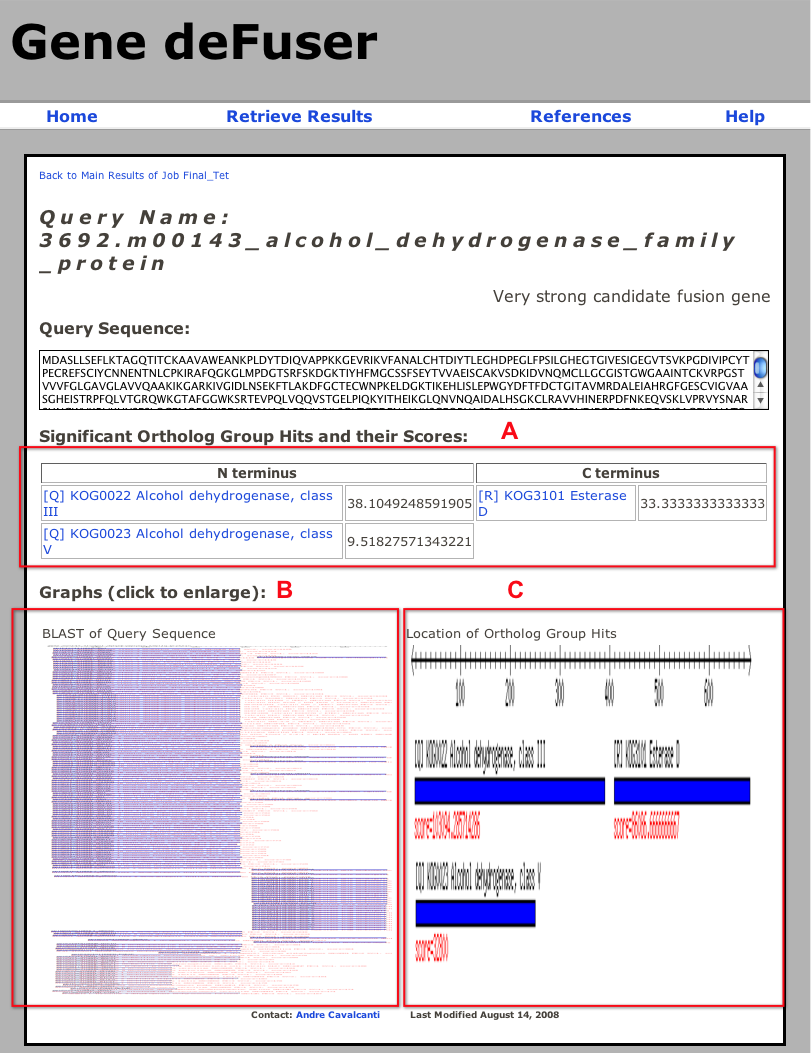
A) Table with the COG hits and scores for the N- and C-termini of the protein. The scores are calcualted using the method described in Zhou and Landweber (2007. Nucleic Acids Res. 35:W678-682). Clicking on the COG name will take to that COG webpage in NCBI.
B) Uniprot BLAST search results. This figure shows the graphical results of a BLAST search of the protein against Uniprot. Clicking on the image will take to a full sized version of the figure.
C) COG search results. This figure shows the location of the COG hits along the full protein. The bars representing the COG hits are the result of merging all the hits to that COG.